Science Spotlight
It's Just a Phase: Defining Phases of Primordial Germ Cell Formation
By Sydney Wyatt
Some argue that germ cells are the most important cell in the body because they alone naturally pass on genetic material to the next generation. Thus, 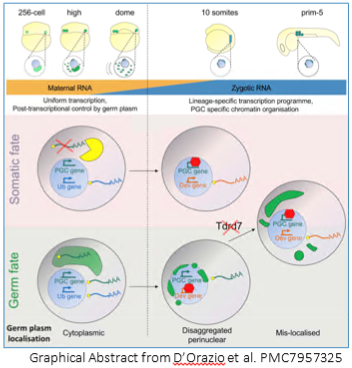 determining what makes a germ cell “decide" to be a germ cell is an important line of inquiry. In sexually reproducing animals, germ fate, as opposed to somatic fate, can be predetermined by oocyte-derived factors contained in the germ plasm, or it can be triggered by the zygotic genome. In zebrafish, the germ plasm is required for germ cell fate determination. However, how early PGCs diverge from somatic cells during their formation beyond the allocation of the required germ plasm remained murky until the recent investigation by Fabio M. D’Orazio et al.
determining what makes a germ cell “decide" to be a germ cell is an important line of inquiry. In sexually reproducing animals, germ fate, as opposed to somatic fate, can be predetermined by oocyte-derived factors contained in the germ plasm, or it can be triggered by the zygotic genome. In zebrafish, the germ plasm is required for germ cell fate determination. However, how early PGCs diverge from somatic cells during their formation beyond the allocation of the required germ plasm remained murky until the recent investigation by Fabio M. D’Orazio et al.
D’Orazio et al used a host of -omics tools — RNA-seq, ATAC-seq and reduced-representation bisulfide sequencing (RRBS-seq) — in conjunction with traditional morpholino knockdown assays and other experiments to examine the profile of PGCs versus somatic cells during early development. They found that PGC formation occurs in two distinct phases bookending zygotic genome activation, with two distinct mechanisms: an early post-transcritptional regulatory phase and a later transcriptional regulatory phase. Before the zygotic genome is activated and before PGC migration begins, germ-plasm-containing cells (future PGCs) have a transcriptome and chromatin accessibility profile similar to somatic cells. Much to their surprise, even germ-cell-specific transcripts were detected in somatic cells. “You don't expect that [PGCs and somatic cells] share a transcriptional program because we are taught that differentiation programs define lineages, but this clearly isn't the case here,” said Ferenc Mueller, the principal investigator on the paper. D’Orazio elaborated that “transcriptional activation is equal throughout the embryo, but it's what happens in the cytoplasm, the shielding of germ-cell-specific transcripts in the germ plasm, that defines the PGCs.”
The second phase of PGC formation occurs after the zygotic genome is active and PGC migration begins. Differential gene expression increased between PGCs and somatic cells, prompting investigation into the epigenome and chromatin accessibility patterns. Unlike mammals in which there is widespread methylation reprogramming, zebrafish PGCs and somatic cells exhibited similar methylation patterns, although upon deeper investigation, PGCs exhibited hypermethylation at putative somatic enhancers. Chromatin accessibility patterns also diverged into PGC- and somatic-cell specific profiles. Interestingly, in PGCs, distal elements became inaccessible while elements proximal to genes promoting germ fate and stem cell differentiation became accessible. The inverse was observed in somatic cells: distal elements were accessible and were enriched for genes promoting developmental and differentiation programs. These changes in gene regulation in PGCs correlated with the time at which the germ plasm relocates from the cytoplasm to a disaggregated perinuclear arrangement, a process which requires Tudor domain 7 (Tdrd7a). The researchers took advantage of this to demonstrate that PGCs without proper germ plasm localization exhibit gene expression and chromatin accessibility patterns more characteristic of somatic cells.
With these new phases come new questions. “As a transcriptional regulation lab, we're interested in following up with factors we identified [in our RNA-seq dataset] which may define the topology organization…in the nucleus,” said Mueller. Experiments investigating their role in transcriptional regulation in PGCs are already underway. “We're also including other datasets like CAGE-seq, which looks at transcription start sites, in our analysis to help investigate our hypothesis that all somatic cells and PGCs share the same transcription machinery,” added D’Orazio. In the far future with some advancements in current technology and information, Mueller said he may be interested in following up with how the germ plasm is physically interacting with the nucleus and the regulatory communication between them.
Reference: D'Orazio, Fabio M et al. “Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization.” Developmental cell vol. 56,5 (2021): 641-656.e5. doi:10.1016/j.devcel.2021.02.007
About the Authors:
 Fabio D’Orazio is a post-doctoral researcher at Imperial College London with Boris Lenhard.
Fabio D’Orazio is a post-doctoral researcher at Imperial College London with Boris Lenhard.
In the before times, he enjoyed traveling around the UK.
 Ferenc Mueller runs a lab at University of Birmingham studying transcriptional regulation at the
Ferenc Mueller runs a lab at University of Birmingham studying transcriptional regulation at the
very first steps of genome activation. His hobby, mountain biking in the forest near his home, was
not affected by COVID but the lack of commute enables him to go almost every day.
 Sydney Wyatt is an Integrative Genetics and Genomics PhD candidate at University of California, Davis, where she works with Dr. Bruce Draper. Currently, she researches early zebrafish gonad development in the context of primordial germ cells and sex determination in the context of the Wnt signaling pathway. You can find her on LinkedIn, Twitter and her website.
Sydney Wyatt is an Integrative Genetics and Genomics PhD candidate at University of California, Davis, where she works with Dr. Bruce Draper. Currently, she researches early zebrafish gonad development in the context of primordial germ cells and sex determination in the context of the Wnt signaling pathway. You can find her on LinkedIn, Twitter and her website.
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.
BLITZing through protein interactions in Zebrafish
By Leo Volkov
To probe the function of a novel protein, it is crucial to identify the interacting partners of the protein of interest. Ideally, one could identify not only strong, direct protein-protein interactions, but also transient and weak interactions. The use of proximity-dependent biotin labeling (BioID) allows one to identify such interactors by making use of biotin ligases fused to a protein of interest. This ligase attaches biotin to all proteins within 10nm. Biotinylated proteins can then be isolated by Streptavidin affinity-based enrichment and identified using various proteomic approaches. However, a fundamental limitation to using this approach in zebrafish has been the need to create transgenic lines in which the protein of interest is directly tagged with the biotin ligase. A novel approach from the group of Thomas E Hall and Robert G Parton, termed BLITZ (Biotin Labelling In Tagged Zebrafish), aims to circumvent this limitation (Xiong et al., 2021) (https://doi.org/10.7554/eLife.64631).
According to the lead author Zherui (Albert) Xiong, the lab was originally motivated to create such a technique to find in vivo protein interactors of Cavin4, a muscle-specific caveola-associated protein. Xiong says “We chose zebrafish because cultured muscle cells do not fully recapitulate the mature muscle fibres in vivo”. To create a modular system for identifying protein interactions, the authors first created a zebrafish line which ubiquitously expresses a biotin ligase (TurboID) fused to a destabilized GFP-binding-peptide (dGBP). When this line is crossed to a zebrafish line that contains a GFP-tagged protein of interest, TurboID-dGBP is recruited to the GFP-tagged protein of interest. Interacting partners near the protein of interest are then biotinylated when biotin is supplemented to the embryo media. These biotinylated proteins can then be isolated and studied for proteomics.
To demonstrate the utility of the BLITZ approach, the authors went on to identify proteins associated with the caveolar cast proteins Cavin1 and Cavin4, in differentiated skeletal muscle. These proteins are crucial for the formation of caveolae, which are specialized invaginations in the cell membrane. TurboID-dGBP fish were crossed with lines that express Cavin1a-Clover, Cavin4a-Clover, and Cavin4b-Clover under the control of muscle specific actin promoter, actc1b. (note: Clover is a GFP recognized by GBP). Liquid chromatography coupled to tandem mass spectrometry (nanoHPLC/MS MS/MS) was then used to analyze biotinylated proteins in embryos that co-expressed the TurboID-dGBP and Cavin-Clover transgenes. Siblings that only expressed the TurboID-dGBP transgene were similarly processed, to serve as background controls. Background proteins, in addition to endogenous biotinylated proteins, and common contaminants were then subtracted. The authors were then able to identify 26, 22, and 25 biotinylated proteins in the Cavin1a-Clover, Cavin4a-Clover, and Cavin4b-Clover samples, respectively.
Consistent with the localization of cavins to the membrane, ~50% of proteins identified were associated with the plasma membrane. Endogenous cavins were also detected in all samples, consistent with the known oligomerization of cavins. The authors also found an ortholog of Pacsin3 (a known caveola-associated protein necessary for muscle caveolar formation) to uniquely interact with Cavin4b. The findings strongly suggest that the modular BLITZ approach is useful for identifying protein-protein interactions in live zebrafish embryos in a cell-type specific manner.
The authors do note a few potential limitations: i) the current indirect method of binding (via GFP, rather than directly to the protein of interest), may expand the labeling radius of the biotin ligase and as a result, detect proteins that do not bind directly to the protein of interest. ii) The binding of GBP to the GFP-tagged protein of interest may also abrogate the binding of certain proteins. iii) The inclusion of a GFP-tag may modify the endogenous binding patterns of the protein of interest. Other tags and validation strategies could be used to address these concerns. Rather than using the bulkier GFP tag fused to the protein of interest, it may be possible to utilize a smaller 15 amino acid biotin acceptor Avi tag in combination with a line in which a biotin ligase is ubiquitously expressed. The Avi tag has previously been used in zebrafish by the Sauka-Spengler lab by creating a transgenic line in which the foxd3 transcription factor was fused to an Avi tag, such that biotinylated foxd3-Avi proteins could then be isolated for chromatin immunoprecipitation (ChIP)-sequencing (Lukoseviciute et al., 2018).
Other concerns can be addressed by the validation of candidate interactors identified the BLITZ approach. Lead author, Zherui (Albert) Xiong, informed us that he is in the process of validating candidate Cavin-interacting proteins by using Förster resonance energy transfer (FRET). With FRET, a protein interaction can be visualized by the FRET emission that occurs when two fluorophore-tagged proteins are within a certain distance of each other. Xiong describes, “We make a construct from our putative list and tag with mScarlet. By injecting mScarlet-tagged candidates into the Clover-Cavin fish, we can validate interactions by colocalization and FRET emission in the fish embryos”. Although the author does note some practical difficulties in cloning larger proteins, the use of such a validation strategy is an exciting next step for the BLITZ approach. In addition, the cavin-associated proteomes discovered in this study highlight the utility and efficacy of the approach. Altogether, the BLITZ technique shows great promise and is sure to expand the world of proteomic studies in zebrafish.
Note: During the writing of this piece, we became aware of another recent publication on the use of biotinylated proximity-based proteomics in zebrafish from the Poss lab (Pronobis et al., 2021). Here, the authors successfully characterized the cardiac proteome during injury and regeneration. We recommend readers take a close look at their study, which shows the potential of using transgenic lines to isolate cell type specific proteomes.
References:
Xiong, Z., Lo, H. P., McMahon, K. A., Martel, N., Jones, A., Hill, M. M., … Hall, T. E. (2021). In vivo proteomic mapping through gfp-directed proximity-dependent biotin labelling in zebrafish. ELife, 10, 1–21. https://doi.org/10.7554/eLife.64631
Lukoseviciute, M., Gavriouchkina, D., Williams, R. M., Hochgreb-Hagele, T., Senanayake, U., Chong-Morrison, V., … Sauka-Spengler, T. (2018). From Pioneer to Repressor: Bimodal foxd3 Activity Dynamically Remodels Neural Crest Regulatory Landscape In Vivo. Developmental Cell, 608–628. https://doi.org/10.1016/j.devcel.2018.11.009
Pronobis, M. I., Zheng, S., Singh, S. P., Goldman, J. A., & Poss, K. D. (2021). In vivo proximity labeling identifies cardiomyocyte protein networks during zebrafish heart regeneration. ELife, 10, 1–22. https://doi.org/10.7554/eLife.66079
About the Technical Report Writer: Leo Volkov is a Neuroscience PhD Student in the laboratory of Joseph Corbo at Washington University, St Louis. Leo work focusses on identifying novel genes that regulate cone subtype specification during retinal development, utilizing single cell RNA sequencing, CRISPR-mediated knockouts, and a variety of other techniques. (Email: leo.volkov@wustl.edu)
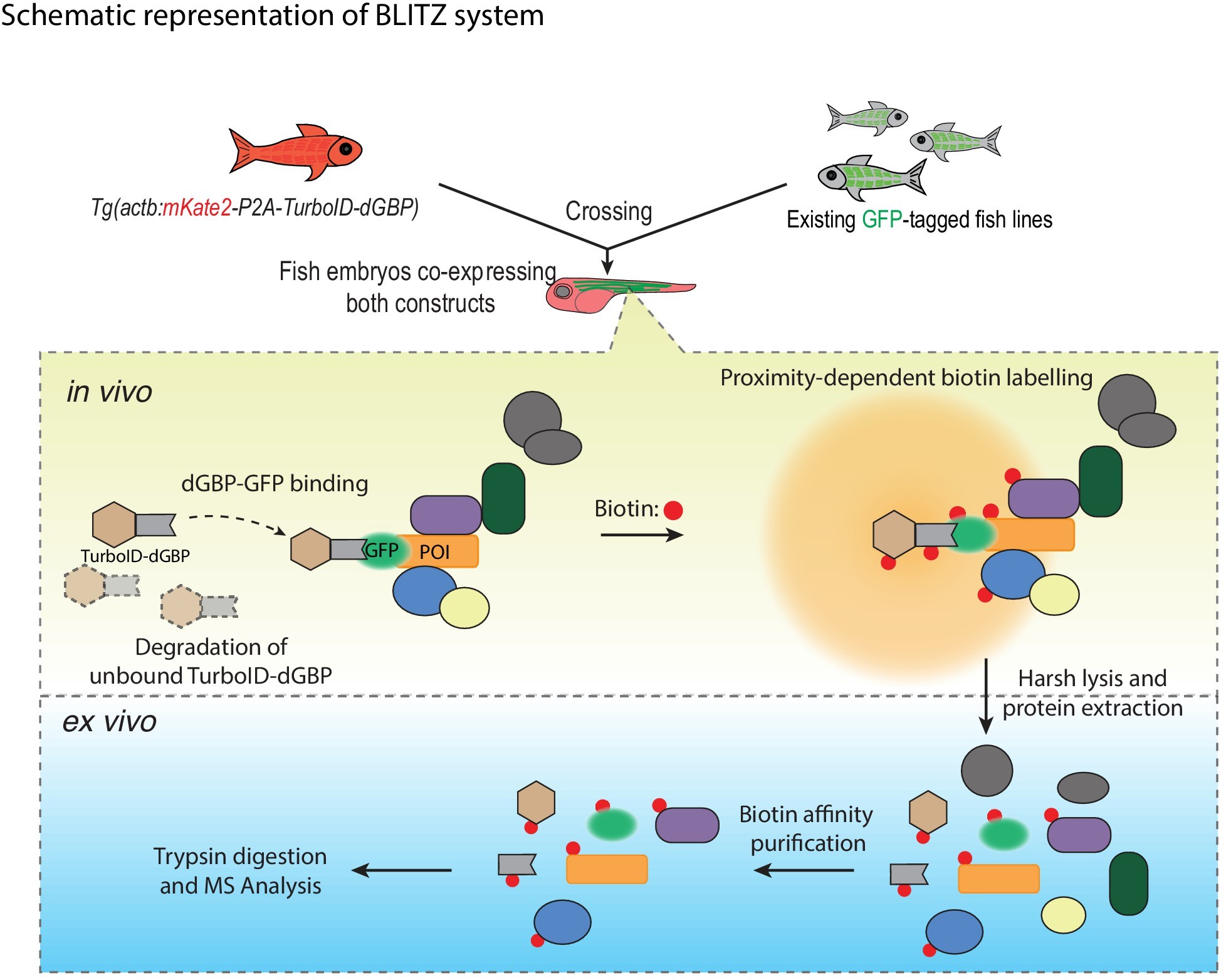
Figure 6 from paper
About the authors of the paper:
Zherui Xiong is a PhD candidate at the Institute of Molecular Bioscience at the University of Queensland in Australia. Thomas E Hall is a senior research officer at the Institute for Molecular Bioscience at the University of Queensland in Australia. Robert G Parton is a group leader at the Institute for Molecular Bioscience and affiliate professor at School of Biomedical Science at the University of Queensland in Australia.

Zherui (Albert) Xiong

Robert G Parton

Thomas E Hall
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.




