Science Spotlight
A New Solution for Sharing Zebrafish Lines
By Dr. Soaleha Shams
Scientific endeavours rely on sharing protocols, reagents and discoveries among researchers. In zebrafish research access to innovative and extraordinary genetic lines can save years of work and money. Re-making transgenic or mutant lines delays progress and the resulting lines are rarely identical to the original due to differences in transgene insertion sites, CRISPR-induced indels and genetic background. While most zebrafish researchers are willing to share lines, the logistics of transporting zebrafish remain challenging, particularly across international borders. Even for domestic transports, researchers may not have space, resources, or permission from their institutions to stock and genotype extra adult fish to send upon request. Shipping embryos is easier, but for the requester it means an extra generation of lost time before imported lines can be used.
To save time and maximize resources, zebrafish sperm can be shipped and used for in vitro fertilization at the destination. However, shipping sperm as it is typically stored—cryopreserved in liquid nitrogen—is problematic given that liquid nitrogen can evaporate during unexpected delays in airports, warehouses, and shipping docks. Furthermore, though sperm cryopreservation is easy using the highly efficient Zebrafish International Resource Center [ZIRC] protocol, in many labs it is still not a standard practice. Fortunately, a solution is at hand: a new report from the Sakai lab at the National Institute of Genetics in Mishima, Japan, may revolutionize the way zebrafish lines are shared by introducing a way of shipping sperm without liquid nitrogen. Takemoto et. al. 2023, reported survival and fertility of zebrafish sperm for four weeks when stored at room temperature in an L-15-based medium supplemented with reagents that are easily available from commercial suppliers. Their novel composition has additional benefits of low cost and minimal effort, and storage without any specialized equipment or extensive procedures. The researchers evaluated the motility and fertility of the stored sperm [F0] after 28 days in this solution and confirmed that the F1 generation from the stored sperm had normal morphology, fertility, and genetic transmission of an EGFP tag to their F2 progeny. Their important discovery that sperm can survive at room temperature in this solution for four weeks can dramatically change how zebrafish lines are shared globally. To prove this point, the authors shipped sperm stored in this medium internationally between labs in Israel, Japan, and the United States without any cooling source and reported successful fertilization at all three destination labs.
Dr. Zoltan Varga, the director of Zebrafish International Resource Center [ZIRC] since 2004, is excited about the publication of the Takemoto et. al. paper. One of ZIRC’s missions is to be a central repository for zebrafish lines. ZIRC imports, cryopreserves, and redistributes genetic lines to the international zebrafish community. For over 20 years, much of this distribution has been in the form of live embryos and adult fish. While zebrafish are quite hardy as a species, they can’t survive the stress of long delays due to extreme weather, holidays, and complicated import regulations. Consequently, international shipments of live fish must be expedited using expensive courier services. Being able to ship sperm labeled as cells rather than live animals, and which can survive inevitable delays, will eliminate this expense.
Of course, shipping sperm rather than fish requires that the receiving lab be able to perform in vitro fertilization. For those unfamiliar with doing IVF, the protocol is in Chapter 2 of The Zebrafish Book, and a video of Charline Walker, who pioneered the approach, is available here (watch between 5:35 and 7:00 in the video). Follow Takemoto et al. for how to fertilize eggs with the shipped sperm.
Shipping sperm may not overcome import regulations regarding potential fish-borne pathogens. The Canadian Food Inspection Agency, for example, bars the import of zebrafish and zebrafish germplasm in its controversial efforts to prevent the transmission of Spring Viraemia of Carp. Nonetheless, Dr. Varga hopes that shipping sperm at ambient temperatures will help zebrafish users in countries without these restrictions reduce cost of domestic and international shipments, and increase flexibility of transport time from a couple of days to almost a month. Though Takemoto et. al. recommend caution while additional research confirms, for example, that sperm stored in their solution is not subject to DNA damage, it is welcome news for those in remote or low-resource settings. ZIRC is currently testing the Takemoto et al protocol for fresh sperm, as well as looking into possibility of thawing cryopreserved samples into the Takemoto solution for room temperature shipping. Varga expects that within a few weeks fish researchers will see updates about options for shipping fresh sperm samples on ZIRC and sister facilities EZRC and CZRC websites soon after.

Set-up for IVF from thawed sperm. Photo taken at the Mayo Clinic zebrafish facility by Soaleha Shams.
About the Science Spotlight Writer
 Soaleha Shams received her PhD in behavioral neuroscience from University of Toronto, Canada, and postdoctoral training in psychopharmacology at Gothenburg University in Sweden. She is currently an NSERC research fellow at the Mayo Clinic in Minnesota, USA, and learning CRISPR-Cas9 gene-editing to manipulate social and stress behaviours in zebrafish. Outside the lab, she uses zebrafish to advocate and work towards improving mental wellness and science literacy in pre-K to grade 12 students through InSciEd Out. She is passionate about reading books with her toddler and YouTube-inspired cooking. She looks forward to starting her own lab in Canada soon to continue studying zebrafish behaviours relevant for better understanding of depression, autism, and stress disorders.
Soaleha Shams received her PhD in behavioral neuroscience from University of Toronto, Canada, and postdoctoral training in psychopharmacology at Gothenburg University in Sweden. She is currently an NSERC research fellow at the Mayo Clinic in Minnesota, USA, and learning CRISPR-Cas9 gene-editing to manipulate social and stress behaviours in zebrafish. Outside the lab, she uses zebrafish to advocate and work towards improving mental wellness and science literacy in pre-K to grade 12 students through InSciEd Out. She is passionate about reading books with her toddler and YouTube-inspired cooking. She looks forward to starting her own lab in Canada soon to continue studying zebrafish behaviours relevant for better understanding of depression, autism, and stress disorders.
Shot through the (silent) heart: Conditional Cardiac Arrest that Gives Degrons a Good Name
By Dr. Timothy Hufford
As scientists, developmental biologists, and zebrafish researchers, we seek a deeper understanding of what occurs within the cells of the organisms surrounding us. Doing so provides key insights as to how our own cells might behave and potentially reveal genes implicated in development, disease, and regeneration. A key strength of organisms such as the zebrafish is their amenability to reverse-genetic approaches. This feature has been leveraged initially through tools such as morpholinos and more recently through the adoption of CRISPR-Cas9 gene editing.
Nonetheless, these tools have some limitations: while morpholinos pose challenges in terms of specificity and toxicity, certain CRISPR alleles have been known to trigger unintended responses such as transcriptional adaptation (El-Brolosy et al., 2019; Sztal & Stainier, 2020). There are also cases in which mutant phenotypes are either embryonic lethal or induce a phenotype at early developmental stages, creating a need for temporally-controlled gene knockouts to study later processes. Conditional knockouts are also ideal when considering stress and recovery conditions, where null mutations may cause death during stress and preclude the study of recovery responses.
Thomas Juan, a post-doctoral researcher in the laboratory of Didier Stanier, set out to recapitulate the tnnt2a/silent heart mutant phenotype by generating a temporal knockout. “Blood flow acts as a mechanical force that instructs endothelial cell development, such as valvulogenesis, and genetic tools to control blood flow have been missing in the field. Here, I provide a means of controlling cardiac contraction in a non-invasive and specific manner.”
Troponin T type 2a (tnnt2a) encodes a thin filament sarcomeric protein required for cardiac contractions, with the aptly named silent heart mutants (Chen et al., 1996; Sehnert et al., 2002) completely lacking heartbeat and displaying major cardiac structural defects stemming from failures in sarcomere assembly. The silent heart mutants are limited by the fact that the lack of heart contractions eventually become lethal, and previous methods to modulate expression such as overexpression, morpholinos, and global recombination lack spatial or temporal specificity.
To develop a Tnnt2a conditional knockout, Juan deployed the Cre-lox recombination approach. Global recombination by injecting Cre mRNA into a floxed tnnt2a allele initially showed promising efficiency. Juan ran into challenges with spatial and temporal recombination, where Cre activity was driven by cardiomyocyte promotors (myl7, myh7, and myh6) either constitutively or through heat shock- or tamoxifen-inducible systems. Rather, the expression using these promotors occurred too late, pre-existing tnnt2a mRNA and Tnnt2a protein masked the phenotype, and the mutant phenotype became apparent only in embryos older than 5 days post-fertilization.
At this point, Juan pivoted to using degrons, which offer a promising and non-invasive means of targeting proteins of interest. The degron (zGRAD) construct contains a GFP nanobody (vhh) to target the protein of interest tagged with GFP, as well as an F-box protein (Nfbxw11b) to promote efficient proteasomal degradation. Testing the Tnnt2a degron under the myl7 cardiomyocyte promoter, Juan found marked reduction of Tnnt2a and successful recapitulation of the tnnt2a mutant phenotype.
Juan also analyzed the Tnnt2a degron embryos at a higher resolution, obtaining single-cell transcriptomes for wildtype, tnnt2a mutants, and Tnn2a degron mutants. UMAP plots revealed an overall striking resemblance between the published mutant and degron transcriptomes, notably diminished populations of epicardial and endothelial cells, as well as a reduction in proliferating cells. However, there were distinct differences between mutant and degron UMAP plots likely reflecting the unique nature of spatial and temporal knockout in the degron line compared to the original mutant line.
After establishing a strong spatial knockout, Juan extended his work and tackled the challenge of gaining temporal control over the Tnnt2a degron by splitting up the vhh nanobody and fusing N-vhh with cpFRB2 and C-vhh with FKBP. The circularly permuted FKBP12 rapamycin-binding domain (cpFRB2) and the FK506-binding protein (FKBP) function as a chemically-induced dimer when in the presence of rapamycin, which acts as a ligand (Lee et al., 2020). In this case, when rapamycin is added and cpFRB2-FKBP form a heterodimer, the N- and C-vhh are joined to form a functional split-zGRAD to target Tnnt2a (see Summary Figure).
Using this split-zGRAD degron led to Tnnt2a degradation in over 50% of the embryos and partially arrested cardiac contraction within 24 hours (see Summary Figure), considerably more rapid than spatially controlled methods involving Cre induction in older embryos. The partial penetrance of the split-zGRAD can possibly be due to factors involving the Tnnt2a already embedded in the sarcomere, the concentration of rapamycin used, or the timing of degradation influenced by dimerization.
The tools established in this work provide compelling evidence in favor of degrons for efficient conditional expression. Juan remarked on the use of degrons as a means of conditional knock-down, “We always have an eye on transcriptional adaptation, and thought this might be a way to avoid this compensation method and to study phenotypes otherwise hidden, there is a lot of potential risk involved.”
As zebrafish mutants and morphants for tnnt2a recapitulate the cardiomyopathy observed in humans harboring TNNT2 mutations, the Tnnt2a degron mutants have strong potential for high-throughput drug-screening and to model this mutation in older zebrafish larvae and adults, which has not been possible through previous methods. The use of degrons in this work not only represents a successful implementation of this technology but offers an intriguing addition to other Cre-mediated recombination strategies. Going forward, degrons also represent a tool which can potentially be applied in combination with other inducible systems such as the Gal4- and Q-systems to modulate expression of multiple proteins of interest in specific cell populations.
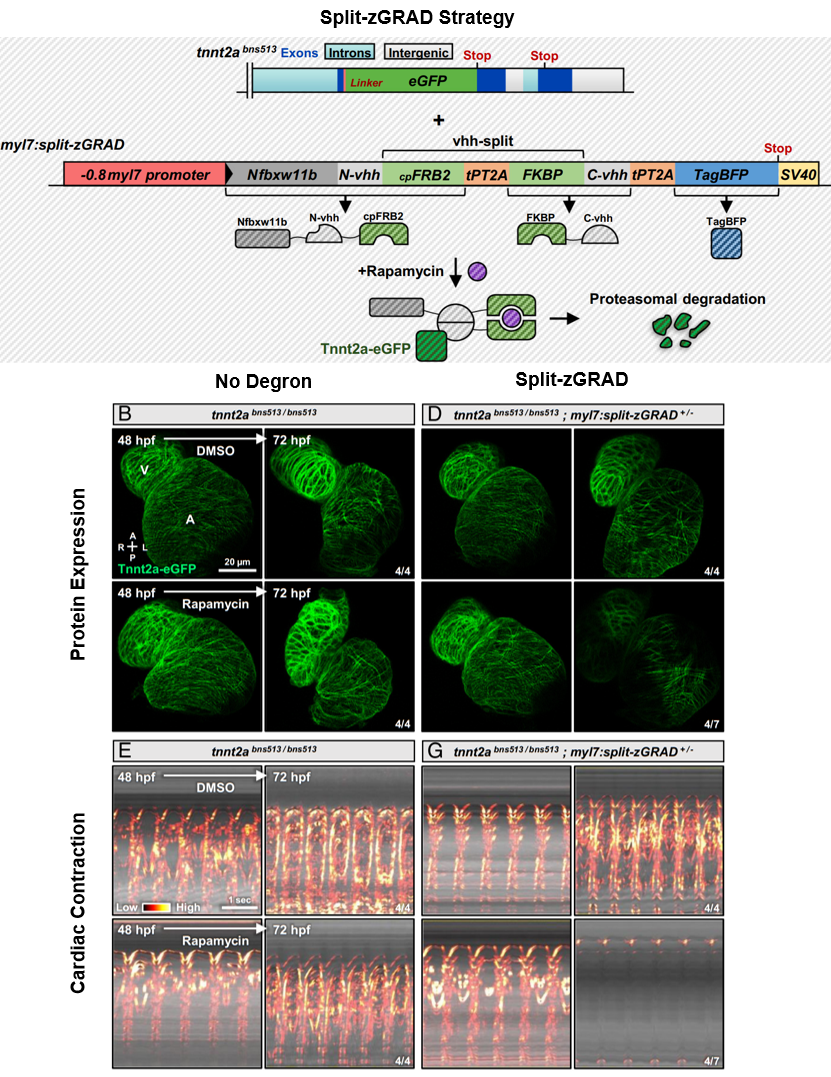
Summary Figure. The degron strategy relies on knocking-in GFP to the target of interest and stably integrating the split-zGRAD construct. The split-GRAD construct (driven by a cardiomyocyte promotor) encodes: 1) Nfbxw11b – an F-box protein to promote degradation of targeted proteins, 2) vhh-split – which, when joined together using the cpFRB2-FKBP system in the presence of rapamycin, form a nanobody that recognizes eGFP, and 3) TagBFP for visualizing split-zGRAD expression. Representative images of Tnnt2a-eGFP expression in the cardiomyocytes of control embryos or embryos expressing the split-zGRAD construct in the presence of DMSO or rapamycin. Kymographs showing the heart contraction rate of the same groups imaged above
References:
Chen, J.-N., Haffter, P., Odenthal, J., Vogelsang, E., Brand, M., Eeden, F. J. M. v., Furutani-Seiki, M., Granato, M., Hammerschmidt, M., Heisenberg, C.-P., Jiang, Y.-J., Kane, D. A., Kelsh, R. N., Mullins, M. C., & Nüsslein-Volhard, C. (1996). Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development, 123(1), 293-302. https://doi.org/10.1242/dev.123.1.293
El-Brolosy, M. A., Kontarakis, Z., Rossi, A., Kuenne, C., Günther, S., Fukuda, N., Kikhi, K., Boezio, G. L. M., Takacs, C. M., Lai, S.-L., Fukuda, R., Gerri, C., Giraldez, A. J., & Stainier, D. Y. R. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature, 568(7751), 193-197. https://doi.org/10.1038/s41586-019-1064-z
Juan, T., Bellec, M., Cardoso, B., Athéa, H., Fukuda, N., Albu, M., Günther, S., Looso, M., & Stainier, D. Y. R. (2024). Control of cardiac contractions using Cre-lox and degron strategies in zebrafish. Proceedings of the National Academy of Sciences, 121(3), e2309842121. https://doi.org/doi:10.1073/pnas.2309842121
Lee, Y. T., He, L., & Zhou, Y. (2020). Expanding the Chemogenetic Toolbox by Circular Permutation. J Mol Biol, 432(10), 3127-3136. https://doi.org/10.1016/j.jmb.2020.03.033
Sehnert, A. J., Huq, A., Weinstein, B. M., Walker, C., Fishman, M., & Stainier, D. Y. R. (2002). Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nature Genetics, 31(1), 106-110. https://doi.org/10.1038/ng875
Sztal, T. E., & Stainier, D. Y. R. (2020). Transcriptional adaptation: a mechanism underlying genetic robustness. Development, 147(15). https://doi.org/10.1242/dev.186452
About the Authors

Dr. Thomas Juan, the first and co-corresponding author of this work, is a Postdoctoral Researcher in Didier Stainier’s laboratory at the Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany. His research interests include cardiac mechanosensation, valvulogenesis, and the creation of genetic tools. In 2023, Thomas was named a PI Fellow as part of Development’s Pathway to Independence. Thomas has recently accepted a research faculty position in the Immunology, Genetics, and Pathology (IGP) department at Uppsala University in Sweden. Please stay tuned for future announcements regarding recruitment. You can also follow Thomas on Twitter at @TJ_ThomasJuan.

Dr. Didier Stainier, a corresponding author of this work, is an Investigator at the Max Planck Institute for Heart and Lung Research in the Department of Developmental Genetics and a leading authority on cardiac development. The Stainier laboratory has been at the forefront of key discoveries including the identification of npas4l as the gene responsible for the cloche phenotype and the elucidation of transcriptional adaptation.
About the Science Spotlight Writer

Dr. Timothy Hufford is a recent PhD graduate from Rachel Brewster’s laboratory at the University of Maryland, Baltimore County. Timothy’s research interests include genetic & cellular adaptations to oxygen deprivation.




