Science Spotlight
Fishing for clues in skeletons: an ancestral gene linked to a recent tuberculosis outbreak.
By Dr. C. M. Santosh Kumar
(Saelens et al (2022) An ancestral mycobacterial effector promotes dissemination of infection. Cell 185 (24), P4507-4525.E18. https://doi.org/10.1016/j.cell.2022.10.019)
This spotlight highlights a recent study published in the journal Cell, where zebrafish were used to unravel a mysterious outbreak of a bone-eating tuberculosis (TB) disease. Tuberculosis (TB) is the most lethal infectious disease caused by the bacterium Mycobacterium tuberculosis (Mtb) and results in about 2M deaths annually, with an additional one third of the world’s population being latently infected (WHO Report 2022).
Although TB is largely a pulmonary infection, a small yet significant proportion (~2%) of cases exhibit a bone disease, known as Pott’s disease (named after a British surgeon, Percivall Pott) or tuberculous spondylitis. Pott’s disease is believed to have existed in 9000-year-old Egypt mummies (Crubezy et al., 1998). Pott’s disease is difficult to detect and treat, and the specific bacterial factors that trigger its extrapulmonary dissemination are not well characterized. The recent TB outbreak that was the subject of this study had Pott’s disease in 57% of the cases, motivating the researchers to search for differences in the mycobacterial genome that might be responsible. They identified a full-length variant of the EsxM gene, a component of the bacterial secretory system required for virulence. This full-length variant is present in ancient Mtb clades while a truncated form of EsxM is present in modern forms of the pathogen. The authors confirmed through a meta-analysis of existing clinical data that the ancient clade is associated with higher levels of Pott’s disease.
Different species of the Mycobacterial genus cause tuberculosis-like infections in animals. For example, M. marinum causes TB-like infections in fish, and M. bovis in cattle. Owing to the speed and ease of use, the Zebrafish M. marinum infection model has emerged as a powerful tool to understand the molecular mechanisms of Mycobacterial infections. Mycobacteria infect host macrophages and cause them to build protective shells (known as granulomas) where the bacteria can survive latently and later disseminate around the host’s body. The dynamics of granuloma formation are thought to influence Mycocbacterial establishment and dissemination. Further, the molecular details of how Mycobacteria tune phagocyte migration and in turn enhance their own fitness and dissemination is not fully understood.
In the study, which the lead author David Tobin described as “really highly collaborative’, the authors employed a multiprong approach, including genomics, proteomics, RNAseq, phylogenetics, infection studies in mice, zebrafish and cell lines to dissect the role of the EsxM in the dissemination of the pathogenic strains. The authors investigated zebrafish – M. marinum infection system for two reasons: i) M. marinum encodes a full-length EsxM that is 87% identical to the ancestral Mtb strain’s full-length EsxM and shares orthologous ESX loci and is overexpressed at comparable levels during infection; ii) The key signalling pathways in bone physiology are conserved between humans and zebrafish. To mimic the modern truncated EsxM variant, the authors introduced a stop codon in M. marinum EsxM. They infected larval zebrafish, and through intravital time lapse imaging, they found M. marinum expressing the full length EsxM had two-fold higher dissemination of infected macrophages from the site of infection compared to M. marinum expressing the truncated form. This effect of EsxM is autonomous to the infected macrophages because macrophages engineered to express full-length EsxM alone exhibited faster migration to wound sites. Further, the authors investigated macrophage migration to the spinal region in juvenile zebrafish. M. marinum expressing full-length esxM had higher rates of bone dissemination, with altered osteoblasts and osteoclast behavior, compared to the strain expressing the truncated variant (Fig. 1). These findings in zebrafish, combined with similar effects on the migration of infected mouse and human macrophage cell lines in culture, allowed the authors to conclude that the full-length EsxM is both necessary and sufficient to induce macrophage migration.
For future directions, Prof. David Tobin, the lead author, says, “Getting a broader understanding of how dissemination can happen is important in understanding and treating the disease”.
The study demonstrates that the form of EsxM in ancient strains of TB influences the macrophages to disseminate more efficiently to the bones than the truncated variant in modern TB, which agrees with the enhanced Pott’s disease observed in humans infected with the ancestral strain. The authors propose that limiting Mtb’s extrapulmonary dissemination may constrain its transmission. Overall, this elegant study of ancient pathogenic strains has identified a novel function for EsxM , which can help better understanding of the patho-adaptation of Mtb and could help develop effective treatment regimens for TB.
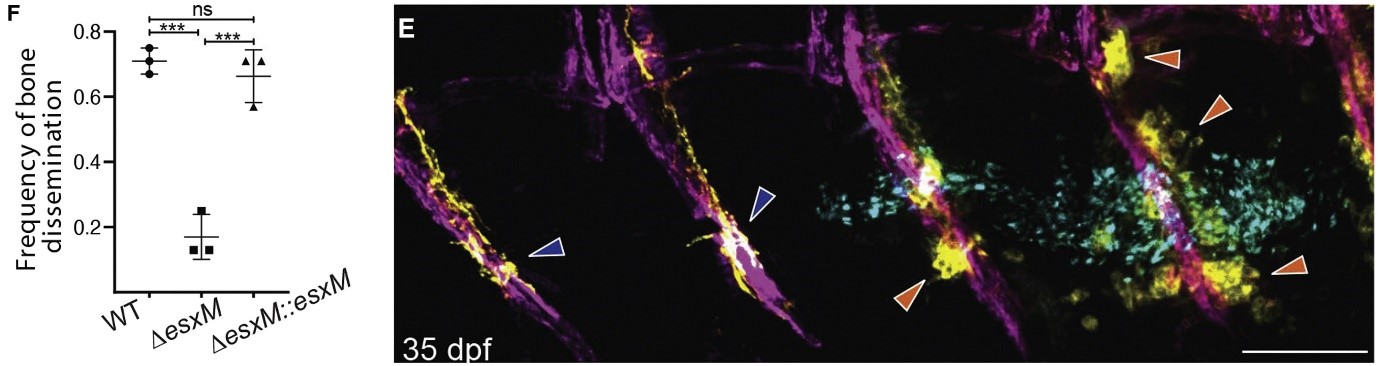
Figure 1. EsxM plays a key role in the bone dissemination. Left: A truncated form of EsxM lowered bone dissemination (and infection), and this was reverted upon complementation with full-length EsxM, indicating a key role for EsxM in dissemination of Mycobacterial infection. Right: Fluorescent microscopic images of cleared infected juvenile zebrafish depict the extent of spinal disease. The affected (with altered osteoclast behavior) and unaffected vertebrae (typical osteoclast behaviour) are shown with orange and dark blue arrowheads, respectively.
References
World Health Organization. Global Tuberculosis Report 2022. World Health Organization, (2022) https://www.who.int/publications/i/item/9789240061729
Crubezy E., Ludes B., Poveda J.D., Clayton J., Crouau-Roy B., Montagnon D. Identification of Mycobacterium DNA in an Egyptian Pott's disease of 5, 400 years old. Comptes rendus de l'Academie des sciences. Serie III, Sciences de la vie. 1998; 321: 941-951 https://doi.org/10.1016/s0764-4469(99)80009-2
J.W. Saelens, M.I. Sweeney, G. Viswanathan, A.M. Xet-Mull, K.L. Jurcic Smith, D.M. Sisk, D.D. Hu, R.M. Cronin, E.J. Hughes, W.J. Brewer, et al. An ancestral mycobacterial effector promotes dissemination of infection. Cell, 185 (2022), pp. 4507-4525.e18 https://doi.org/10.1016/j.cell.2022.10.019
About the Authors
 Joseph W. Saelens, Mollie I. Sweeney and Gopinath Viswanathan are the joint first authors of this article. Joseph initiated this work as a PhD student. Joe is now working as a Computational Biologist at Pfizer Inc., Cambridge, USA. The work was continued by Mollie Sweeney and Gopinath Viswanathan. Mollie is a PhD student in the lab and is interested in understanding microbiological and molecular genetic features of Mycobacterial infections using zebrafish. Gopinathan is a postdoc in the Tobin lab and is interested in understanding host response to Mycobacterial infection in zebrafish.
Joseph W. Saelens, Mollie I. Sweeney and Gopinath Viswanathan are the joint first authors of this article. Joseph initiated this work as a PhD student. Joe is now working as a Computational Biologist at Pfizer Inc., Cambridge, USA. The work was continued by Mollie Sweeney and Gopinath Viswanathan. Mollie is a PhD student in the lab and is interested in understanding microbiological and molecular genetic features of Mycobacterial infections using zebrafish. Gopinathan is a postdoc in the Tobin lab and is interested in understanding host response to Mycobacterial infection in zebrafish.
Authors’ group photo: Left to right, Mollie Sweeney, Ana María Xet Mull, David Tobin, Gopinath Viswanathan.
Prof. David M. Tobin, the senior author of the paper, is a Professor of Molecular Genetics and Microbiology at the Duke University School of Medicine, USA. His research is focussed on understanding Mycobacterial pathogenesis and host susceptibility.
 Joseph W. Saelens
Joseph W. Saelens
About the News Splash Science Writer

Dr. C. M. Santosh Kumar is a Research Fellow with the group of Prof. Gurdyal Besra at the University of Birmingham, Birmingham, UK. Santosh established the Zebrafish M. marinum infection model at Birmingham. His long-standing research interest has been to develop novel therapeutic regimens to tuberculosis.
Finding Your Way Through the Zebrafish Brain
By Vindhya Chaganty
The development of neural circuitry can be actively mapped using dynamic expression of genes that act as molecular landmarks of neuronal subpopulations. Additionally, the function of neurons can depend on their connectivity to other neurons and behavioural stimuli. This makes it important to have visualisation tools that can integrate multiple modalities allowing us to correlate gene expression, circuit connectivity and functional activity.
One substantial contribution was the Allen Brain atlas which allowed visualisation of thousands of genes in both mouse [1] and human brain sections [2]. However, mapping whole brain at a cellular resolution is difficult in brains with larger size and higher complexity. Additionally, using brains sections does not allow for mapping neuronal activity in response to real-world stimuli. Whole brain atlases and connectomes have been established in non-vertebrate model organisms like Drosophila melanogaster [3] with much simpler nervous systems and are valuable databases to study neuronal cell fate, function, and connectivity.
The zebrafish is being used extensively to approach questions in neuroscience, making it important to understand its neural circuitry. Herwig Baier’s group at the Max Planck Institute for Biological Intelligence near Munich, Germany, previously generated an integrated tool that provided subcellular resolution to the neuroanatomy of 6 day old (dpf) larval zebrafish brains [4]. At this stage, larvae already exhibit some complex behaviours and, owing to their near transparency, allow high resolution whole brain imaging. The Max Planck Zebrafish Brain or “mapZebrain” atlas is an integrated multimodal database of transgenic lines, histological data and single neuron morphological reconstructions. However, this atlas was missing a corresponding spatial map of gene expression at a cellular resolution. Co-first author Dr. Inbal Shainer explained, “Working with RNA sequencing data, you want to know where the genes are expressed. But available resources are limited in developmental timeline, lack 3D resolution and it is tough to merge in-situs from multiple genes. So, the need for integrated FISH data was always there”.
Now, Shainer et al. (2023) have enriched the MapZebrain atlas by integrating high-resolution RNA in situ images. Their tool enables researchers to classify neurons using multiple features with high spatial accuracy on the larval zebrafish brain. Shainer et al., used multiplexed fluorescent in-situ RNA hybridisation chain reaction (HCR) technique [5] to label 290 target genes previously established as cell type specific markers, many with important functions in cell fate-specification, neuromodulation and neurotransmission. They acquired images at cellular resolution using volumetric confocal imaging and used computational co-registration of the images onto a reference brain. The data is now available on mapZebrain.org which allows users to search neurons based on brain region and markers. Since this FISH data is integrated with the previous mapZebrain datasets, users can investigate spatial overlaps of their gene of interest with other features as well. Dr. Shainer briefly mentioned that they are still adding a few genes at a time to the dataset with some more genes currently in progress.
Apart from being a discovery and identification tool, the authors were also able to exhibit the power of this atlas to help identify behaviourally relevant neural circuitry. They exposed larvae to several sensory and behavioural paradigms that are commonly studied. Following this, they used an unbiased whole brain activity mapping by post-hoc FISH staining for cfos (fosab, which is a marker for recent sustained neuronal activity). This led to the identification of a hindbrain population - the secondary gustatory nucleus (SGN), which was previously unknown in the context of prey capture. Using the multimodality of this integrated atlas, they further characterised the gene expression and projection patterns of SGN neurons to other regions of the brain to identify a novel pathway related to prey capture. For continuous improvement of the multimodality of this atlas, the authors aim to “find a way to bridge this with calcium imaging data to see specific neuronal activity with temporal resolution which is missing with cfos”.
Despite being a powerful tool, this dataset comes with a few limitations. Firstly, it focuses on an early larval stage which may not be ideal to study complex behaviours such as social interactions, learning memory etc. Additionally, despite efforts by the authors to enhance visualisation of deeper ventral brain regions, gene expression information could be lost in these regions.
With this multimodal, user-friendly web tool, researchers can visualise, co-register their own data, and download relevant data. Dr. Shainer stressed that this project was made possible by a group effort including “co-first author Enrico Kuehn, Eva Laurell who made the single cell tracings that were added to the atlas and Mariam-Al-Kassar who is in-charge of the website”. Baier’s group is currently using this atlas to “look at spatial organisation of cell types in particular brain regions”. It is also being used to “better define the anatomy of the zebrafish brain”. Overall, this paper is a crucial addition to the growing resources for zebrafish neuroscience research.
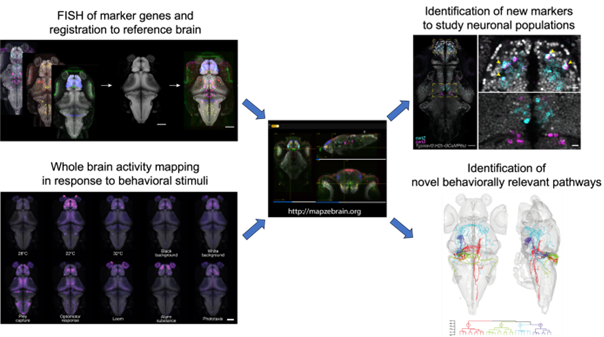
Fig. adapted from Shainer et al., 2023. A graphical summary illustrating the workflow involved in developing this atlas (left) and its potential applications (right). The marker gene expression patterns and post-hoc activity mapping (using cfos HCR) were registered onto a reference brain and integrated into “mapZebrain”. This data was then accessed to examine overlap of interesting markers that can be used to label neurons of interest. The activity maps were used to identify novel clusters which can be characterised using the multimodal approaches of the atlas.
Source:
Inbal Shainer et al. , A single-cell resolution gene expression atlas of the larval zebrafish brain.Sci. Adv (2023) doi:10.1126/sciadv.ade9909
References:
1. Lein, E.S., et al., Genome-wide atlas of gene expression in the adult mouse brain. Nature, 2007. 445(7124): p. 168-76.
2. Hawrylycz, M.J., et al., An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 2012. 489(7416): p. 391-399.
3. Milyaev, N., et al., The Virtual Fly Brain browser and query interface. Bioinformatics, 2012. 28(3): p. 411-5
4. Kunst, M., et al., A Cellular-Resolution Atlas of the Larval Zebrafish Brain. Neuron, 2019. 103(1): p. 21-38 e5.
5. Choi, H.M.T., et al., Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development, 2018. 145(12).
About the Authors

Co-first author Inbal Shainer is a postdoc in Herwig Baier’s group at the Max Planck Institute for Biological Intelligence where she continues to work on the zebrafish brain atlas. She is also interested in understanding the spatial expression of genes in the optic tectum.

Co-first author Enrico Kuehn is a molecular biologist in Herwig Baier’s group at the Max Planck Institute for Biological Intelligence. He is a part of the “Brain atlas” team and continues to work on improving the cellular resolution atlas of the zebrafish brain.

Herwig Baier is the director of the Max Planck Institute for Biological Intelligence and the head of the “Genes-Circuits-Behaviour” department. The research focus of his group is to understand how sensory inputs and internal state information are integrated by neuronal circuits and converted into behavioural responses.
About the News Splash Science Writer
 Vindhya Chaganty is a research fellow in Dr. Caroline Wee’s group at the Institute of Molecular and Cell Biology, A*STAR in Singapore. Her current research focus is understanding the neural circuitry and genetics of appetite regulation using zebrafish and mouse models.
Vindhya Chaganty is a research fellow in Dr. Caroline Wee’s group at the Institute of Molecular and Cell Biology, A*STAR in Singapore. Her current research focus is understanding the neural circuitry and genetics of appetite regulation using zebrafish and mouse models.




