Science Spotlight
Planes, Trains, and autoinhibition domains: kinesin specialization regulates the behavior of distinct neuronal compartments
By Adam J Isabella
Locomotion is a fundamental aspect of our lives. Even the laziest among us can rarely go more than a few hours without moving about, and society is ceaselessly enchanted with the invention of new devices to overcome our inherent locomotory limitations, from the simple – roller skates, bikes, electric scooters – to the incredibly complex – cars, trains, planes, spaceships. So fundamental is locomotion, in fact, that it does much more than just get us from point A to point B. For seemingly however far you zoom in on the workings of the human body, there you will find it. Examine our cells, and you will find immune cells marching through the body in search of foreign invaders and skin cells crawling to cover a wound. Zoom even further, to the very molecules that compose these cells, and you will find bustling actin and microtubule tracks upon which trains of motor proteins transport cargo.
Most, if not all, cellular functions rely on microtubule transport, which is driven largely by Kinesin and Dynein motors, and as with all things in the cell, this system must be highly organized. Dr. Mary Halloran, professor at the University of Wisconsin Madison, highlights a central paradox of such a ubiquitous system: “The Kinesin-1 motor can transport so many [cargos], but how does one motor deliver such diverse cargos to specific cell locations?” Consider, for example, a favorite cell of the Halloran lab – the Rohon-Beard (RB) sensory neuron. “One of the most interesting things about the RB sensory neurons is that they have these central axons and peripheral axons that act very differently...and the cell has to template them differently,” explains postdoctoral fellow Dr. Liz Haynes. In a new article in eLife, Drs. Haynes and Halloran, in collaboration with the labs of Drs. Jan Huisken and Kevin Eliceiri, identify a key role for a Kinesin-1 component in specifying RB axon domains.
Kinesin-1 is a polymer of kinesin heavy chain (KHC) and kinesin light chain (KLC) proteins. While KHCs have been more heavily studied, KLCs stood out to the group as more likely confer specificity in cargo transport because, in addition to their well-known role in regulating Kinesin-1 autoinhibition, these proteins mediate cargo binding. Their diversity – vertebrates have four klc genes – also suggested the potential for specialization. To understand how KLCs impact RB organization and function, the authors knocked out the RB-expressed klc4 gene. The impacts they observed were clear but subtle, suggesting the mutant does not broadly disrupt Kinesin-based trafficking. Using live imaging to characterize axon behavior, the authors did not note any change in central axon behavior, but reported two peripheral axon phenotypes – first, while these axons normally branch extensively, mutants had fewer branches, resulting from a defect in stabilizing newly formed branches. This defect is, in turn, likely due to disrupted microtubule dynamics, as the group observed increased microtubule dynamicity and reduced polymerization of microtubules into nascent branches. Second, while peripheral axons normally repel each other on contact, axons from mutants instead tended to fasciculate together. Importantly, both of these mutant behaviors are more reminiscent of the normal function of RB central axons, supporting the idea that KLC4 may transport cargos to peripheral axons to support their specific functionality. Finally, the authors asked whether the cellular defects of klc4 loss of function impact the animal’s behavior. Indeed, they found that mutant larvae are hypersensitive to touch (likely a direct result of sensory neuron defects), and that larval and adult fish show increased anxiety, suggesting a broader role for KLC4 in neural circuit development.
Reflecting on the impact of this work, “one of the contributions our paper made is to have people think a little bit differently about the roles of KLC’s, that they aren’t just needed for maintenance of the axon or neuron or for bulk transport of materials, but that they actually have specific roles in patterning a developing neuron and making different parts of the cell different from one another,” explained Dr. Halloran. Looking forward, Dr. Halloran is excited to identify the KLC4 cargos that are responsible for promoting appropriate peripheral axon behaviors. Dr. Haynes, meanwhile, plans to move into imaging neurons in the adult brain to identify the cause of the behavioral defects in these fish: “What I would really like to do, ultimately in my own lab…is to go from the developmental phenotype, what is this single gene doing during development, link that to what’s going on in the larva, and in the adult...using non-invasive [2-photon] imaging.” Dr. Haynes was also eager to point out the value of the zebrafish klc4 mutant as a disease model, as klc4 mutations in humans can cause the neurodegenerative disease hereditary spastic paraplegia, but the neurological cause of this disease is not known.
About the Authors
 Dr. Elizabeth “Liz” Haynes was a postdoctoral fellow in Dr. Halloran’s lab at the University of Wisconsin Madison from 2015-2021, and is now a Morgridge Post-doctoral fellow with Drs. Kevin Eliceiri and Tyler Ulland at the Morgridge Institute for Research. She is interested in live imaging and in understanding the genetic basis of neurodevelopmental and neurodegenerative disease. More info can be found at www.emhaynes.com/
Dr. Elizabeth “Liz” Haynes was a postdoctoral fellow in Dr. Halloran’s lab at the University of Wisconsin Madison from 2015-2021, and is now a Morgridge Post-doctoral fellow with Drs. Kevin Eliceiri and Tyler Ulland at the Morgridge Institute for Research. She is interested in live imaging and in understanding the genetic basis of neurodevelopmental and neurodegenerative disease. More info can be found at www.emhaynes.com/
 Dr. Mary Halloran is a Professor in the Department of Integrative Biology at the University of Wisconsin Madison. Her lab is interested in understanding the mechanisms that control axon growth and guidance. More info can be found at https://integrativebiology.wisc.edu/staff/halloran-mary/
Dr. Mary Halloran is a Professor in the Department of Integrative Biology at the University of Wisconsin Madison. Her lab is interested in understanding the mechanisms that control axon growth and guidance. More info can be found at https://integrativebiology.wisc.edu/staff/halloran-mary/
 Dr. Adam Isabella is a postdoctoral fellow in Dr. Cecilia Moens’s lab at the Fred Hutchinson Cancer Center and an incoming assistant professor of Genetics, Cell Biology, and Development at the University of Minnesota. He is interested in the patterning of neural circuitry during development and regeneration. More info can be found at www.isabella-lab.org
Dr. Adam Isabella is a postdoctoral fellow in Dr. Cecilia Moens’s lab at the Fred Hutchinson Cancer Center and an incoming assistant professor of Genetics, Cell Biology, and Development at the University of Minnesota. He is interested in the patterning of neural circuitry during development and regeneration. More info can be found at www.isabella-lab.org
Haynes EM, Burnett KH, He J, Jean-Pierre MW, Jarzyna M, Eliceiri KW, Huisken J, Halloran MC. KLC4 shapes axon arbors during development and mediates adult behavior. Elife. 2022 Oct 12;11:e74270. doi: 10.7554/eLife.74270. PMID: 36222498; PMCID: PMC9596160.
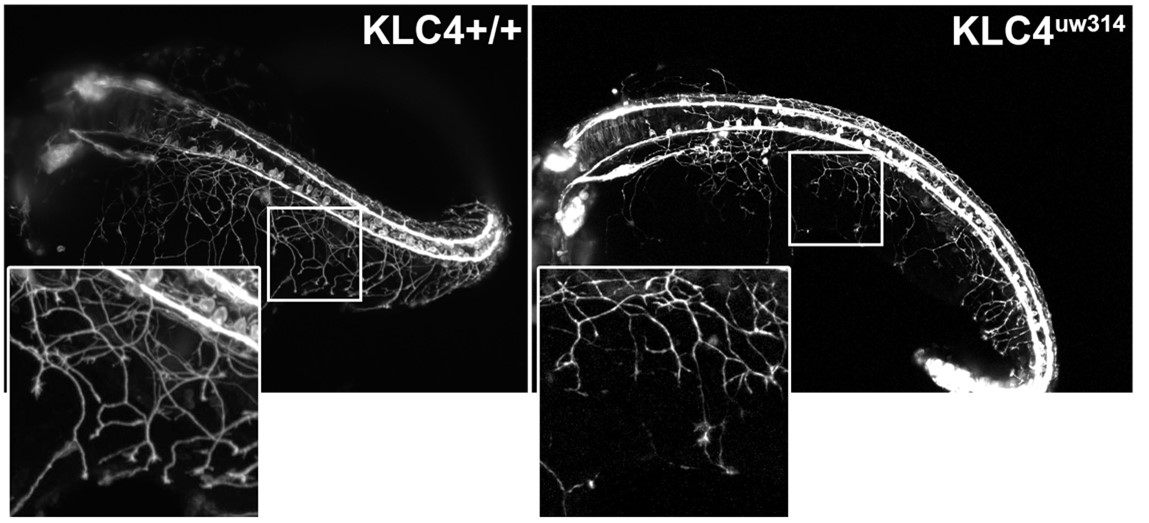
Caption: RB neurons in wild-type (left) and klc4 mutant (right) zebrafish at 24hpf showing decreased branching and coverage of peripheral axons in the mutant. Image provide by Dr. Liz Haynes.
Seeing is believing, but seeing bigger is better!
By Clyde Pinto
The amenability of zebrafish to be imaged both fixed and live has revealed many fundamental processes and the basis of disease states. However, the ability to distinguish between two closely spaced particles, is limited by the resolution of a conventional light microscopy which is around 200-300 nm for instance, when imaging GFP. While it is easy to resolve large structures such as cells and even nuclei, clarity is lost when examining smaller subcellular structures. Approaches with better resolution will be useful for studying nano scale structures such as centrosomes, nuclear pore complexes or even small cellular protrusions such as cilia and microvilli. Many ‘super-resolution’ microscopy techniques have been developed in recent years, but these for the most part, require specialized equipment that many labs cannot easily access. A recent approach, Expansion microscopy, turns the paradigm on its head by changing the sample instead of the microscope (Chen et al., 2015).
Expansion microscopy uses special gel chemistries to embed samples which can then be hydrated to expand the sample equally in all directions. The samples expand approximately 4-fold to even 10-fold and can be imaged on regular microscopes. This leads to an improvement in resolution to ~50 nm. The gel compositions are simple to use for standard experimental laboratories and reagents are available commercially but the approach needs to be optimized for each type of sample to ensure appropriate expansion.
There are broadly two approaches to expansion microscopy: 1) conventional expansion microscopy (ExM), in which the sample is first stained with fluorophores that are then crosslinked to the gel, followed by proteinase K treatment to breakdown the proteins and hydration of the gel resulting in expansion (Wassie et al., 2019). 2) In a second approach, the sample is fixed to the gel followed by denaturation using SDS and heat. The sample can then be stained after partial expansion of the gel. The latter approach is called magnified analysis of the proteome or MAP (Ku et al., 2016). MAP methods have the advantage that staining partially expanded samples potentially allows antibody penetration into otherwise densely packed organelles that resist staining, and in addition, decrease in the linkage error (the error induced as a consequence of the size of the antibody), which further scales on expansion. Ultrastructure-expansion microscopy (U-ExM) is a newly-developed modification of the MAP method which ensures preservation of cellular ultrastructure (Gambrotto et al., 2019). While ExM has previously been used to image larval zebrafish brains and more recently to the whole larvae (Freifeld et al., 2017; Sim et al., 2021), U-ExM has only been applied to isolated molecules, single cells and selected tissues.
A new paper in Cell Reports Methods from the Vermot lab at Imperial College London and their collaborators at University College London and MRC Harwell Institute provides the first standardized protocol for expansion microscopy of whole zebrafish larvae with a modification of the MAP approach, that retains tissue ultrastructure (Steib et al., 2022). The authors named this approach TissUExM and showed that in addition to zebrafish, the methodology works with whole mouse embryos and Drosophila wing imaginal discs as well. The first author, Emmanuelle Steib, had prior experience with U-ExM from her Ph.D. work and Julien Vermot the principal investigator encouraged Emmanuelle to apply this to zebrafish to help understand the biology of the cilia in a high resolution, molecular and tissue context, which is difficult with other methods such as electron microscopy.
However, expanding zebrafish came with its challenges due to “the size and the mechanical properties of the sample” said Emmanuelle. Their initial experiments with 2dpf larvae resulted in cracking of the samples during expansion. They reasoned that some of the steps of the protocol needed optimization. Addition of a detergent (0.1% triton-X100) ensured that the reagents penetrated uniformly into the sample. The authors then modified the kinetics of gelation, incubation temperatures and times to ensure homogenous sample gelation, loss of intra-sample resistance and preservation of epitopes. Together, these improvements allowed successful isotropic expansion of 2dpf larvae, with concomitant maintenance of morphology and cellular ultrastructure as well as epitope preservation for immunofluorescence, and the authors were able to study cilia structure in various tissues.
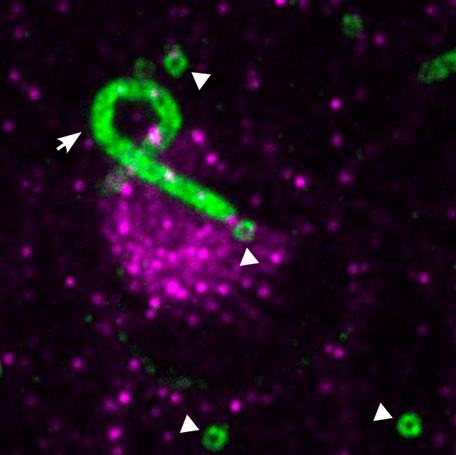
Caption: TissUExM of the inner ear of a 2dpf zebrafish embryo, with centrioles (white arrowheads) and a kinocilium(white arrow) in green, surrounded by stereocilia in magenta. Image Credits: Modified from an image by Emmanuelle Steib, Julien Vermot Lab.
A potential concern with the modified approach is that as zebrafish larvae age their collagen network becomes more resistant to expansion. This was especially evident from 3 dpf. So, in order to expand older larvae, Steib and coworkers included a collagenase VII treatment prior to denaturation. This allowed proper expansion of the sample. Additionally, they were also able to show uniform expansion was possible using commonly employed fixatives for zebrafish, PFA and Dent’s.
Emmanuelle had some advice for those trying expansion microscopy. She said “it takes time and practice” and one needs to be “patient” as “it might not work the first time”. This is because it is a 2-week long process, and mistakes in any of the critical steps require a restart from the beginning. “However, once it’s established it’s extremely reproducible and extremely robust”.
About the Authors

Emmanuelle Steib is a postdoc in the Vermot lab at Imperial College London, focussing on the structure and functions of primary cilia during cardiovascular development. She has been awarded a Marie Sklodowska Curie Individual Fellowship to establish a novel super-resolution method for high precision imaging in whole zebrafish embryos. Her aim is to provide a comprehensive and dynamic description of primary cilia characteristics from organelle- to tissue-scale.
Emmanuelle initially studied Pharmaceutical Sciences at the University of Strasbourg (France), then obtained a PhD in Life Sciences from the University of Geneva (Switzerland).
 Julien Vermot leads the biomechanics and signalling lab focusing on the understanding on the impact of mechanical stresses during morphogenetic and regenerative processes.Julien obtained his PhD in developmental biology from the University of Strasbourg in 2003, where he worked on the role of retinoic acid during embryonic development. He then worked as a visiting scientist the Stowers Institute for Biomedical Research in Kansas City, USA, followed by a post-doctoral position at the California Institute of Technology in Pasadena where he developed new tools to study the role of mechanical forces during development. He was Research Director at the French INSERM before joining the Department of Bioengineering at Imperial College London in 2019.For his research, he has won numerous prizes, including selection for the EMBO Young Investigator program (YIP), the Career Development Award from the HFSP and the ERC Consolidator Grant.
Julien Vermot leads the biomechanics and signalling lab focusing on the understanding on the impact of mechanical stresses during morphogenetic and regenerative processes.Julien obtained his PhD in developmental biology from the University of Strasbourg in 2003, where he worked on the role of retinoic acid during embryonic development. He then worked as a visiting scientist the Stowers Institute for Biomedical Research in Kansas City, USA, followed by a post-doctoral position at the California Institute of Technology in Pasadena where he developed new tools to study the role of mechanical forces during development. He was Research Director at the French INSERM before joining the Department of Bioengineering at Imperial College London in 2019.For his research, he has won numerous prizes, including selection for the EMBO Young Investigator program (YIP), the Career Development Award from the HFSP and the ERC Consolidator Grant.
 Clyde Pinto is a post-doctoral research fellow at the University of Warwick, UK. He studied actin based protrusions in the skin and intestine of zebrafish during his PhD at the Tata Institute of Fundamental Research, India. During his post-doctoral fellowship he studied modifications of the actin cytoskeleton in zebrafish and yeast in the labs of Karuna Sampath, Mohan Balasubramanian and Masanori Mishima funded by the BBSRC and currently works to uncover the molecular mechanisms of cytokinesis in eukaryotes funded by the University of Warwick.
Clyde Pinto is a post-doctoral research fellow at the University of Warwick, UK. He studied actin based protrusions in the skin and intestine of zebrafish during his PhD at the Tata Institute of Fundamental Research, India. During his post-doctoral fellowship he studied modifications of the actin cytoskeleton in zebrafish and yeast in the labs of Karuna Sampath, Mohan Balasubramanian and Masanori Mishima funded by the BBSRC and currently works to uncover the molecular mechanisms of cytokinesis in eukaryotes funded by the University of Warwick.
References
Chen, F., Tillberg, P. W., & Boyden, E. S. (2015). Expansion microscopy. Science, 347(6221), 543-548. https://pubmed.ncbi.nlm.nih.gov/25592419/
Freifeld, L., Odstrcil, I., Förster, D., Ramirez, A., Gagnon, J. A., Randlett, O., ... & Boyden, E. S. (2017). Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proceedings of the National Academy of Sciences, 114(50), E10799-E10808. https://pubmed.ncbi.nlm.nih.gov/29162696/
Gambarotto, D., Zwettler, F. U., Le Guennec, M., Schmidt-Cernohorska, M., Fortun, D., Borgers, S., ... & Guichard, P. (2019). Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nature methods, 16(1), 71-74. https://pubmed.ncbi.nlm.nih.gov/30559430/
Ku, T., Swaney, J., Park, J. Y., Albanese, A., Murray, E., Cho, J. H., ... & Chung, K. (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nature biotechnology, 34(9), 973-981. https://pubmed.ncbi.nlm.nih.gov/27454740/
Sim, J., Park, C. E., Cho, I., Min, K., Lee, J. S., Chong, Y., ... & Chang, J. B. (2021). Whole-ExM: expansion microscopy imaging of all anatomical structures of whole larval zebrafish. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.05.18.443629v1
Steib, E., Tetley, R., Laine, R. F., Norris, D. P., Mao, Y., & Vermot, J. (2022). TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy. Cell reports methods, 2(10), 100311. https://pubmed.ncbi.nlm.nih.gov/36313808/
Wassie, A. T., Zhao, Y., & Boyden, E. S. (2019). Expansion microscopy: principles and uses in biological research. Nature methods, 16(1), 33-41. https://pubmed.ncbi.nlm.nih.gov/30573813/https://doi.org/10.1038/s41592-018-0219-4
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.




