Science Spotlight
Flipping the Metabolic Switch for Tail Fin Regeneration
By Susanna Riley
Mention the regeneration of a limb, and most people will consider it as something from science fiction, but much research has in fact focussed on this process. One animal that can regenerate complex appendages is the zebrafish, which responds to tail amputation with the formation of a blastema, a mass of undifferentiated highly proliferative cells capable of replacing all lost cell types. Mammals typically have limited capacity to form a blastema, and this likely underlies their inability to regenerate appendages. Much work has focussed on elucidating the triggers and maintenance of the blastema but understanding how cells dedifferentiate and proliferate to become the fin blastema is incomplete. In recent work published in npj Regenerative Medicine, Sinclair et al. sought to elucidate the role of metabolism, often altered in rapidly proliferating and differentiating cells, in this process.
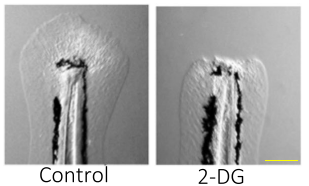
After amputation of the embryonic tail fin, this paper demonstrates that a metabolic shift is essential for the formation of a functional blastema. “Even in the 70’s people had made assertions that perhaps there was metabolic switching during regeneration” says first author Dr. Jason Sinclair. “[The] lab had done microarray on regenerating zebrafish tails and some of the genes that came out of it had to do with glycolysis”. This was supported by findings that 2-DG administration, which inhibits glycolysis, resulted in a complete failure of regeneration. Single-cell RNA sequencing of the regenerating tail fin with and without 2-DG showed that 2-DG altered mesenchyme gene expression, inhibiting the formation of a blastema but not affecting the development of the uninjured fin-fold. Interestingly, genes involved in TGF-β signalling were downregulated by 2-DG. The TGF-β family of growth factors has prominent roles in a range of cellular processes and many diseases, including cancer. Pharmacological inhibition of TGF-β signalling phenocopied 2-DG treatment and blocked regeneration, suggesting that metabolic reprogramming is required for TGF-β signalling.
Sinclair et al. also used CRISPR/Cas9 to elucidate which metabolic pathway triggers regeneration. Only genetic perturbation of the hexosamine biosynthetic pathway (HBP) by gfpt1/2 loss phenocopied 2-DG treatment, with mutants being hypersensitive to 2-DG. “I think the one that got us really excited, and when we knew we had it, was when we had a gene-drug interaction” says Sinclair. “If you've not got [gfpt1/2] you could use 10 times less of the drug and still get the same effect, which really shows it's the pathway that matters” adds PI Dr. Shawn Burgess. Normally, HBP utilises glycolytic intermediates to generate charged sugars for glycosylation. Together, this paper posits that regeneration after tail fin injury requires a metabolic shift to increase glucose flux through the HBP. This promotes N-linked glycosylation, which triggers signalling pathways such as TGF- β, required for the formation of normal blastema mesenchyme.
This finding suggests that a Warburg-like physiology is associated with tail fin regeneration. The Warburg effect is more commonly associated with cancer biology. Characterised by increased glucose uptake and lactate production, it allows cancerous cells to promote survival and proliferation without relying on aerobic respiration. “It made some sense to us that there may be a similar metabolic switch during regeneration that there is in cancer because the processes are very similar” says Sinclair. Both regeneration and cancer have similar transcriptional profiles and require cell proliferation, increased mesenchyme formation, and, as investigated in this study, a shift to glucose-dependent metabolism. This raises difficulties in the treatment of both cancer and regeneration-related disorders. For example, chemotherapy targeting highly proliferative cancerous cells also kills proliferative regenerating cells, such as the hair follicle. Therefore, investigation of the similarities and differences between regeneration and cancer may provide mechanistic insight into how to design carefully targeted drug treatments that can manipulate one without affecting the other.
This work could also be linked to regeneration in other organisms. “We're super interested in seeing that the phenomenon is basic and conserved across evolution” says Burgess. “We suspect it is, of course, but we'd like to show that that this is an ancient phenomenon that recurs over and over again anytime a blastema forms during regeneration”. Mammalian blastemas can be found in limited situations, including digit tip regeneration in children and African spiny mice. This suggests that a better understanding of the blastema could help to promote its formation in creatures that otherwise have a limited regenerative capacity. It may even bring us closer to the elusive whole limb regeneration seen in science fiction, although hopefully without the dubious scientific explanations and catastrophic side effects!
Dr. Jason Sinclair (first author) did the work that this paper covers as a postdoc in the Burgess lab. He currently works at Sirnaomics. Outside of science he likes to play bass guitar and work out.
Dr. Shawn Burgess (PI) runs his lab at the NIH, focussing on developing efficient gene knockout and phenotyping technologies in the zebrafish. Pre-pandemic he loved travelling but in recent times he enjoys discovering his local hiking trails.
Susanna Riley (author of this article) is a Wellcome funded Tissue Repair PhD student in the Gram Hansen and Feng labs at the University of Edinburgh, where she studies the Hippo pathway in zebrafish regeneration. When not in the lab, she likes playing D&D and testing out new recipes.
Paper reference: “The Warburg effect is necessary to promote glycosylation in the blastema during zebrafish tail regeneration” PMID: 34518542, DOI: 10.1038/s41536-021-00163-x
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.
Space Jam, Space SLAM: Uncovering spatio-temporal transcriptomics of single-cell zebrafish embryos
By Vicky Tan
Gene expression programs drive the spatial organisation of various cell types during embryonic development. The intracellular localisation of maternal transcripts is crucial to many embryonic developmental processes, including animal–vegetal (AV) axis formation1. Current approaches such as single-cell RNA sequencing (scRNA-seq), allow us to follow differentiation trajectories of cells during development, but do not provide information about the sub-cellular location or how RNAs are transported. Spatial resolution of transcriptomics of single cells is important for understanding the dynamics of developmental processes and how it may ultimately influence cell fate and function. Recent methods including Multiplexed error-robust fluorescent in situ hybridisation (MERFISH) and other microscopy-based techniques are powerful in discerning spatial transcriptomics at single-cell resolution, however, is difficult to apply this to embryos at all stages. Therefore, there is a need for methods that can simultaneously resolve both spatial and temporal RNA expression at the single-cell level. Building on previous work, Holler et al. (2021) have developed a method for elucidating the spatio-temporal transcriptome of the first few hours of zebrafish development2.
To study the RNA spatial patterns of a one-cell stage zebrafish embryo, Holler et al. applied tomo-seq. By barcoding and sequencing 96 sections along the AV axis of the embryo, they identified known localisation patterns of some vegetally-localised genes that are important for patterning (dazl, trim36), and validated their distribution by whole mount in situ hybridisation (WISH)3,4. Vegetally-localised genes contribute to germ cell development and dorsoventral axis specification, and showed the most reproducible spatial patterning within the one-cell embryo. The authors then focussed on genes localised at the embryonic vegetal pole and built a repository of 97 genes expressed in this region. This is a ten-fold increase from previously known vegetal genes, and highlights the utility of the tomo-seq method in identifying novel RNA localisation patterns.
To distinguish between maternal and zygotic transcripts, the authors then used a single-cell RNA metabolic labelling method known as ‘scSLAM-seq’. A nucleotide analogue, 4-thiouridine (4sU) was injected into single-cell embryos, which were then harvested during gastrulation (6 hpf), when both maternal and zygotic transcripts are present. This allowed Holler et al. to identify uridine-labelled transcripts as the 4sU is incorporated into newly transcribed zygotic transcripts. Cells localised in the enveloping layer had the highest labelling rate, indicating a high transcriptional or proliferative rate among zygotic cells. In contrast, zygotic contribution to primordial germ cells (PGCs) appeared lower, reflected by lower labelling efficiency in these cells. This is consistent with previous literature showing that PGCs are specified by inheritance of maternal determinants5,6. Amongst the 97 vegetally-localised genes Holler et al identified, 47 were unlabelled (i.e., maternally-derived) and highly expressed. Maternally-derived vegetal transcripts clustered into two subpopulations of PGCs after gastrulation. The authors identified novel candidates and tracked their fates within these different populations, to determine their role in germ cell specification and differentiation.
Holler et al. then investigated the conservation of germ cell factors of vegetally-localised genes between zebrafish and Xenopus. Tomo-seq was applied to two Xenopus species (X.laevis and X.tropicalis) and identified some known localisation patterns. Nine vegetally-localised genes were robustly found in all three species including previously known factors dazl and syntabulin, highlighting the ability of this method to identify RNA localisation at subcellular resolution in a range of organisms. The 3’ untranslated regions (UTRs) of many RNAs have cis-regulatory localisation elements that are thought to drive transcript localisation. However, 3’UTR elements have not been well studied in zebrafish. Holler et al. identified 216 isoforms of vegetally-localised genes in zebrafish and found that their 3’UTR sequences were 1.7-fold longer than background. Whilst the functional importance of longer 3’UTR sequences remains unclear, the group hypothesise that the UTRs might harbour elements that could be important for cellular localisation, cytoskeletal anchoring and RNA stability. Lastly, the group found the same polyU-motif in vegetally localised genes across both Xenopus species and zebrafish and propose that they may have important conserved roles in maternal RNA stability.
Holler et al. elegantly integrated their highly sensitive tomo-seq and scSLAM-seq methods to understand the dynamics and fate of transcripts important for patterning along the embryonic AV axis. The authors acknowledge that sectioning from single cells smaller than zebrafish embryos (~700 µM) may be difficult. Thus, the combination of tomo-seq and scSLAM-seq will require further optimisation for in vivo studies in small samples or for studying organ systems, as 4sU delivery and incorporation is likely to differ between tissues. Nevertheless, the group were able to distinguish maternal and zygotic transcripts, which can aid understanding of how they localise and contribute to cell fates in developing embryos. The authors also showed that the shared regulatory elements in the 3’UTR of different genes between zebrafish and Xenopus may provide insights into RNA localisation mechanisms.
The first author Karoline Holler emphasised the importance of having proper controls and taking time to optimise techniques such as tomo-seq and scSLAM-seq. She also encourages scientists to have fun to stay motivated. Holler is now applying these techniques to better understand olfactory receptors. The senior author of this paper, Philipp Junker, said that scSLAM-seq may not be as straight forward for certain cell types or tissues, where variable 4sU labelling rates or differences in metabolism between different cells might need to be considered. This can be controlled for by looking at uridine incorporation with and without perturbation. Computational methods and mathematical modelling can also assist in accounting for other factors that may affect the data, such as differing mRNA decay rates.
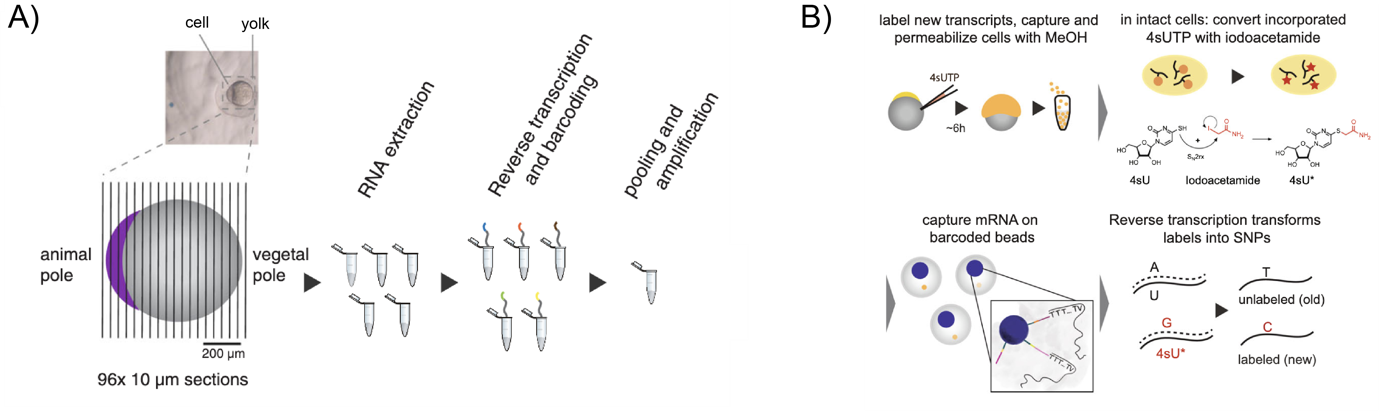
Figure legend: A) Schematic of single-cell sectioning for tomo-seq. B) Schematic of 4sU labelling at single-cell stage and methanol fixation of cell to permeabilise membrane for nucleotide analogue conversion by iodoacetamide. Single cells are barcoded and labelled nascent zygotic transcripts can be identified.
About the Authors
Dr. Karoline Holler is a post-doctoral researcher with a passion for computational biology and genomics at the Helmholtz Centre Munich. She completed this highlighted work during her PhD at the Max Delbrück Centre in Berlin. Her current work focusses on physics and data-based modelling of cellular decision making.
Dr. Jan Philipp Junker is the head of the Quantitative Developmental Biology laboratory at Max Delbrück Centre in Berlin. His laboratory has a strong interest in understanding embryonic development by combining biophysics, systems biology, and developmental biology.

About the Technical Report Writer
Vicky Tan is currently completing her PhD under the joint supervision of Dr. Andrew Cox and Professor Mark Dawson at the Peter MacCallum Cancer Institute in Melbourne. Her research involves developing new tools to better understand immediate transcriptional responses following acute liver injury and during liver regeneration.

Highlighted Paper
Holler, K. et al. Spatio-temporal mRNA tracking in the early zebrafish embryo. Nature Communications 12, 3358, doi:10.1038/s41467-021-23834-1 (2021).
Additional References
- Escobar-Aguirre, M., Elkouby, Y. M. & Mullins, M. C. in Vertebrate Development: Maternal to Zygotic Control (eds Francisco Pelegri, Michael Danilchik, & Ann Sutherland) 173-207 (Springer International Publishing, 2017).
- Holler, K. et al. Spatio-temporal mRNA tracking in the early zebrafish embryo. Nature Communications 12, 3358, doi:10.1038/s41467-021-23834-1 (2021).
- Langdon, Y. G. & Mullins, M. C. Maternal and Zygotic Control of Zebrafish Dorsoventral Axial Patterning. Annual Review of Genetics 45, 357-377, doi:10.1146/annurev-genet-110410-132517 (2011).
- Welch, E. & Pelegri, F. Cortical depth and differential transport of vegetally localized dorsal and germ line determinants in the zebrafish embryo. BioArchitecture 5, 13-26, doi:10.1080/19490992.2015.1080891 (2015).
- Raz, E. Primordial germ-cell development: the zebrafish perspective. Nature Reviews Genetics 4, 690-700, doi:10.1038/nrg1154 (2003).
- Yoon, C., Kawakami, K. & Hopkins, N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124, 3157-3165 (1997).
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.




