Science Spotlight
A Direct Route to Spinal Cord Repair
By Özgün Özalp
Spinal cord injury in humans is a life-altering event that can result in permanent loss of sensation and movement. A goal of regenerative medicine is to improve outcomes by activating cells at the site of injury to regenerate neurons and neuronal processes to bridge the damage and reconnect the brain with the body below the site of the injury. But which cells are to be reactivated, and by which signals? To answer these questions, scientists have turned to animals like zebrafish that regenerate easily, with the goal of translating discoveries made there into humans. In work recently published in Developmental Cell, Cavone et al. discover a direct role for macrophages in signaling to neural progenitors to initiate neurogenesis after spinal cord injury.
Spinal cord injury in fish and amphibians causes resident neural stem cells, called ependymo-radial glia (ERG), to initiate neurogenesis. Macrophages are known to promote this regenerative neurogenesis, however the underlying mechanisms are not known. “It was not clear at the beginning which cells communicate with what type of cells” says co-P.I. of the study Thomas Becker. Furthermore, the mechanism is unlikely to be the same as that which controls neurogenesis during embryogenesis. “Cells at the injury site are exposed to signals they may not have seen during development, yet they are able to re-initiate neurogenesis. That means they must have the receptors to respond to the new injury signals and our progenitor cells may be in an epigenetic state that predisposes them to neurogenesis” explains co-P.I. Catherina Becker. In this study, Cavone et al. investigated the hypothesis that specific pro-regenerative macrophages signal directly to progenitor cells, causing the re-initiation of neurogenesis. Since macrophages play no role in normal developmental neurogenesis, this article has shed light into regeneration-specific signaling pathways driving neurogenesis.
The first hint that macrophages might signal directly to neural progenitors came from single-cell RNA sequencing, which identified a transcriptional signature of tumor necrosis factor (TNF) signaling within ERG progenitors FACs-purified after spinal cord injury (TNF is produced predominantly by macrophages). The investigators went on to show that pharmacological or genetic suppression of TNF signaling suppressed neurogenesis, while treatment of ERG progenitors ex vivo with TNF triggered them to initiate neurogenesis. To demonstrate a progenitor-autonomous requirement for TNF signaling in neurogenesis the authors used the Tet-ON system to express a dominant-negative form of Hdac1 in progenitors at the time of injury. Hdac1 is an epigenetic regulator of motor neuron differentiation whose expression in ERGs is induced by TNF signaling and whose activity promotes regenerative neurogenesis. Excitingly, the authors found that Hdac1 inhibition specifically in ERGs suppressed neurogenesis. Asked what was the most challenging aspect of the work, Becker and her PhD student and co-first author Louisa Drake agreed: injuring and dissecting 200 larval fish to isolate enough cells for each scRNA-Seq experiment was hard work!
Taken together, the work strongly supports a model in which pro-regenerative macrophages are recruited to the site of injury where they directly induce neural progenitors to initiate neurogenesis via TNF signaling. However, the effects of TNF on regeneration were partial, suggesting that parallel regenerative mechanisms are at work that have yet to be discovered. The experiments were done in larval fish for reasons of accessibility and convenience, but does the direct signaling mechanism continue to drive spinal cord regeneration adults? Finally, a major issue is that in mammals, TNF worsens recovery from spinal cord injury, likely by promoting inflammation and scarring. Consequently, any novel therapeutic approaches involving TNF that emerge from the work will need to be carefully targeted to neural progenitors.
About the authors
The work was done at the Center for Discovery Brain Sciences at the University of Edinburgh.
Dr. Leonardo Cavone (first author) was a post-doc in the Becker lab who has now moved into industry and was not available for this interview.
Dr. Thomas Becker (Co-PI, Becker Lab) studies gene regulation leading to reprogramming of quiescent ERGs for regenerative neurogenesis, focusing on histone acetylation and immune signals as mediators of the regenerative response.
Professor Catherina Becker (Co-PI, Becker Lab): has pioneered the use of zebrafish to study spinal cord regeneration. Her research team studies the factors governing generation of neurons and axonal pathfinding in the CNS during development and regeneration, using the zebrafish model to identify fundamental mechanisms in vertebrates with clear translational implications for CNS injury and neurodegenerative diseases.
Özgün Özalp, the author of this Science Spotlight article, is a PhD student in the Ozhan Lab at the Izmir Biomedicine and Genome Center (IBG) at Dokuz Eylul University Health Campus in Izmir, TURKEY.
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.
Knock Knock Knock-in of Fluorophores: A New Method of Endogenous Terminal Tagging
By Evan Brown
Transgenic lines expressing fluorescent proteins are essential tools for zebrafish research. With such lines, we can observe cellular behaviors, detect changes in signaling, visualize proteins, and much more. Current approaches to generating these lines, however, have inherent limitations especially when investigating protein function and dynamics. Injection of mRNA, random integration of transgenes, and bacterial artificial chromosomes (BAC) often do not recapitulate endogenous expression patterns of the protein of interest. Faithful expression patterns—in space, time, and level—are necessary to probe the physiological and functional properties of a protein. The best way to achieve fidelity is by tagging the gene via a knock-in approach, creating a fusion protein at the endogenous locus. Yet, generating knock-ins has its own difficulties: it utilizes inefficient DNA repair pathways (e.g. low rate of homologous recombination compared to non-homologous end joining) and can introduce indels around the insertion site, resulting in out-of-frame tagged proteins that fail to fluoresce. Consequently, the zebrafish community lacks an extensive suite of transgenic lines with knocked-in fluorophore tags. Levic et al. (2021) seeks to solve this problem with a new method for endogenous protein tagging.
Levic et al. developed two similar, but slightly different methods for C- and N-terminal endogenous protein tagging to get around the aforementioned issues. Both approaches rely on CRISPR-Cas9 with gRNA that cuts in an intron and injecting a linearized, dsDNA donor amplicon. For the C-terminus, the amplicon contains a homology arm that spans part of the last intron and the entire last exon, the fluorophore coding sequence, and a polyadenylation sequence. The N-terminus relies on a 5’ homology arm upstream and through the 5’UTR, the fluorophore coding sequence, and a 3’ homology arm that spans the first exon and part of the first intron. After the Cas9-induced DNA break, the amplicon integrates into the genome through non-homologous end joining. Because the break and insertion occur in a non-coding region, which is removed from transcripts via RNA splicing, mutations and imprecise integrations are much less likely to affect expression. Indeed, Levic et al. successfully tagged the C- or N-terminus of several genes with this method, demonstrating stable and efficient germline transmission for both large and small fluorophore tags.
To test how faithfully endogenous expression patterns were recapitulated upon tagging, the authors generated a suite of C- and N-terminus knock-in tags for various epithelial genes. For example, different fluorophore knock-ins for ZO-1 (ZO-1-tdTomato and ZO-1-eGFP) perfectly colocalized in epithelial cells. Cldn15la-tdTomato, when compared to an established cldn15la BAC transgene, showed very similar expression patterns and unlike the BAC (Alvers et al., 2014), was homozygous viable. The authors also generated Itgb1b-tdTomato to visualize cell-ECM adhesions, eGFP-Rab11a to detect membrane trafficking, and eGFP-aPKC to distinguish the apical polarity complex. Levic et al. then used these knock-ins to measure the relative abundance of labeled proteins in a cell. By using the signal decay profile of eGFP molecules, they inferred and compared the abundance of Rab11a on apical vesicles of eGFP-rab11a knock-in fish. Despite differences in rab11a expression, lysosome-rich enterocytes (LRE) and intestinal epithelial cells (IEC) had similar concentrations of Rab11a on apical vesicles. However, in LREs, they found a greater proportion of vesicles with more than three eGFP molecules, suggesting that a subset of vesicles may specialize in protein uptake. Their result suggests a potentially novel mechanism by which LREs internalize dietary proteins in the gut.
Although this method greatly improves upon existing strategies, there are a few salient limitations. First, the exogenous polyA tail in the C-terminus amplicon may change mRNA stability; this worry can be resolved by substituting the endogenous gene’s polyA tail sequence in the amplicon. Second, it relies on the accuracy of CRISPR-Cas9 to cut only in a non-coding region and not in nearby exon sequences. Third, if the targeted intronic or other non-coding region encompass miRNA target sites or other regulatory elements, the tagging approach might disrupt gene expression. Lastly, like many transgenic methods, this method is a somewhat time-consuming process: about 5% of injected F0 fish show mosaic expression of the knock-in and depending on the strength of the fluorophore signal, F0s may require confocal imaging to screen. Nonetheless, with this new knock-in technique, zebrafish researchers have an improved means of endogenously tagging the C- or N-terminus of a protein. Regarding endogenous fusions, Levic says, “The [zebrafish] field [has] had a lot of hurdles compared to other models.” With this development, the community will hopefully see an increase in new knock-in lines that will facilitate the study of protein dynamics throughout development and across cell types. He adds, “I hope that [this method] will encourage others to maybe invest a little bit of time to try and make the best tools that they can so that the field as a whole can put our best foot forward and do the best work that we can.”
Reference: Levic, D.S., Yamaguchi, N., Wang, S., Knaut, H. and Bagnat, M. (2021). Knock-in tagging in zebrafish facilitated by insertion into non-coding regions. Development 148(19): dev199994. https://doi.org/10.1242/dev.199994.
About the Authors:
Dan Levic is a postdoctoral researcher at Duke University in the Bagnat Lab. He received a B.S. from Middle Tennessee State University and completed his Ph.D. in Cell and Developmental Biology at Vanderbilt University School of Medicine. Dan studies membrane protein trafficking/sorting in the intestine.
Michel Bagnat, the principal investigator, studied for his PhD in Germany at the European Molecular Biology Laboratory (Heidelberg) and the Max Planck Institute (Dresden). He then moved to the United States, first to work as a postdoc in Didier Stainier’s lab at the University of California, San Francisco, and started his lab at Duke University in 2008. The Bagnat group’s research focuses on understanding the cellular and physiologic mechanisms controlling morphogenesis, including how hydrostatic pressure influences development.
About the Technical Report Writer:
Evan Brown is a Biology PhD Candidate working in Dr. Sarah Kucenas’ lab at the University of Virginia in Charlottesville, VA. Evan’s research focuses on the development and function of satellite glial cells in dorsal root ganglia, their cellular interactions, and contributions to somatosensory dysfunction.
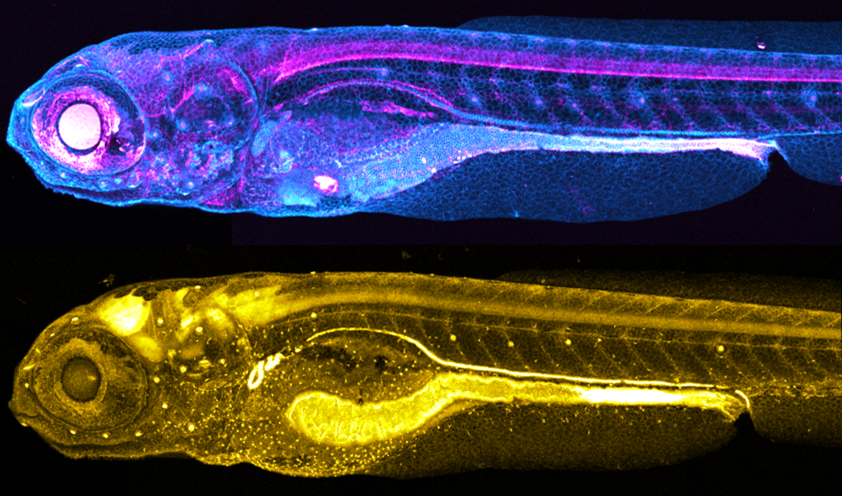
Transgenic knock-in fish generated by the Levic et al. method. (Top) TgKI(tjp1a-tdTomato) at 7 dpf; (Bottom) TgKI(eGFP-rab11a) at 5 dpf.
Would you like to have a mentored science writing experience as a Science Spotlight author for NewsSplash? If you are interested, please contact Cecilia Moens (cmoens@fredhutch.org) or Karuna Sampath (K.Sampath@warwick.ac.uk) for more details about joining our team.




